Improved Artificial Ribosome More Efficient
New tethered ribosome design increases efficiency of protein expression for synthetic biology
Four years ago, Northwestern Engineering professor Michael Jewett created the first artificial ribosome, leading the way to use cells to create new kinds of therapeutics and polymers.
But the ribosome, called Ribo-T, faced several challenges: it caused cells to grow slowly, and generated lower protein expression than the cell usually generated.
Now, Jewett, professor of chemical and biological engineering and co-director of the Center of Synthetic Biology, and his collaborators are back with a new and improved version, which increases the efficiency of Ribo-T’s protein expression. The new system provides a better platform for researchers to study this cornerstone of biology.
 “This new version of Ribo-T will allow us and other labs around the world to jumpstart the synthesis and evolution of new therapeutics and materials made from engineered ribosomes,” Jewett said.
“This new version of Ribo-T will allow us and other labs around the world to jumpstart the synthesis and evolution of new therapeutics and materials made from engineered ribosomes,” Jewett said.
The results were published September 2 in the journal Nature Communications. The work was done in collaboration with researchers at the University of Illinois at Chicago.
Creating two operating systems
Ribosomes are the engines of cells: they make all the cell’s proteins, which carry out the cell’s functions. Biochemical engineers have harnessed this power to transform the lives of millions of people through the synthesis of biopharmaceuticals, like insulin, and industrial enzymes, like subtilisin, that are used in laundry detergents. Synthetic biologists have looked to harness this power to direct ribosomes to create new kinds of proteins, peptidomimetic drugs that address rising antibiotic resistance, and polymers that could lead to next-generation materials, like stronger, lighter, and sensor-embedded fabrics.
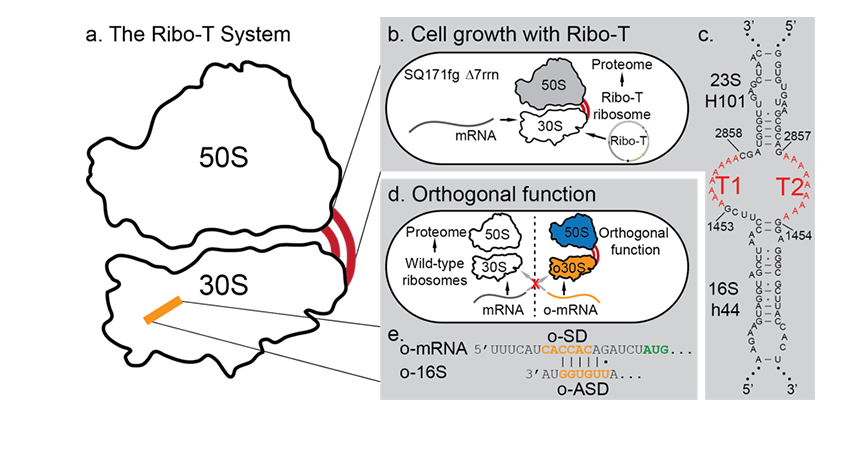
To make a protein, the ribosome’s two subunits — one large and one small — unite on messenger RNA (mRNA). That combined unit assembles the protein before separating back into subunits.
Ribo-T’s major advancement was to permanently link these two subunits together through tethers, creating what was essentially a dual operating system. The “wild-type” part of the ribosome could continue keeping the cell alive, while the second operating system — the artificial ribosome — could be used as the researchers wanted: to make new kinds of proteins the cell would not normally create.
“Ribosomes are found in all kingdoms of life, and until Ribo-T, it was thought that the separate but interacting nature of their subunits was required of successful protein synthesis,” said Jewett, a Charles Deering McCormick Professor of Teaching Excellence. “We showed for the first time that that this assumption was incorrect, which opened a transformational new direction for repurposing ribosomes for synthetic biology.”
Improving tethers, efficiency
Though the system worked, it was difficult to use. The cells remained alive, but did not work as efficiently as before. So for the past several years, the researchers have worked to optimize the system.
At issue was the tethers to the subunits and the base-pairing interaction, which allows the subunits to receive the specialized messages that give them directions on what proteins to create.
The researchers screened hundreds of thousands of new tether sequences to find the best option to tether the two subunits together. They also found the optimal base pairing interactions for messages.
“We were able to evolve a new and improved Ribo-T,” said Anne d’Aquino, a graduate student in Jewett’s lab and coauthor on the paper.
New Ribo-T more efficient
Cells containing the new Ribo-T had a significant increase in growth rate compared to the original version, and had a 208 percent increase in overall expression of the target protein. The researchers also used it to successfully synthesize a range of proteins with different sizes and structures.
The new Ribo-T is much more accessible and user-friendly, and several laboratories are already using it.
“Now that we have this optimized system, we can use it to synthesize new proteins or materials that would have otherwise been too slow and difficult with the old system,” d’Aquino said. “Many researchers are now going to be able to use this platform to engineer better proteins and materials.”