Revealing the Rules Behind Virus Scaffold Construction
New insights into epistasis could impact future drug delivery and therapeutic strategies
A team of researchers including Northwestern Engineering faculty has expanded the understanding of how virus shells self-assemble, an important step toward developing techniques that use viruses as vehicles to deliver targeted drugs and therapeutics throughout the body.
By performing multiple amino acid substitutions, the researchers discovered instances of epistasis, a phenomenon in which two changes produce a behavior different from the behavior that each change causes individually.
 “We found occurrences where two separate single amino acid changes caused the virus shell to break or become really unstable, but making both changes together produced a stable structure that functioned better than ever,” said Danielle Tullman-Ercek, associate professor of chemical and biological engineering at the McCormick School of Engineering.
“We found occurrences where two separate single amino acid changes caused the virus shell to break or become really unstable, but making both changes together produced a stable structure that functioned better than ever,” said Danielle Tullman-Ercek, associate professor of chemical and biological engineering at the McCormick School of Engineering.
The paper titled “Experimental Evaluation of Coevolution in a Self-Assembling Particle,” was published March 19 in the journal Biochemistry. Tullman-Ercek served as the paper’s co-corresponding author along with collaborator Matthew Francis, professor of chemistry at the University of California at Berkeley.
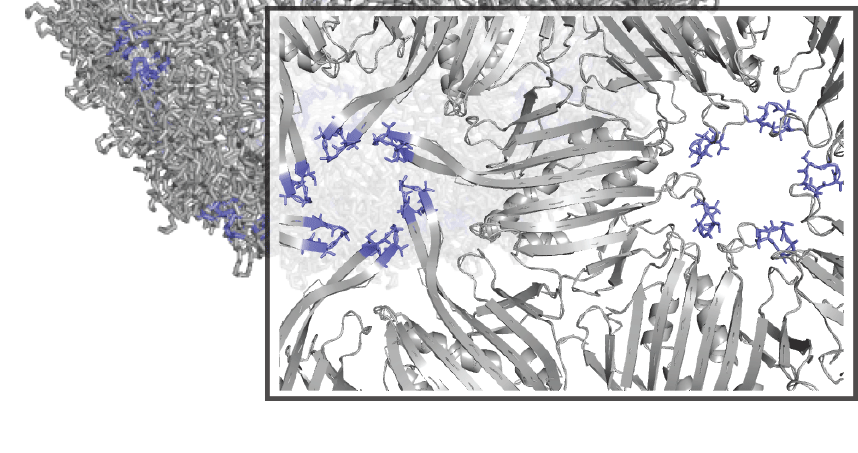
The work builds on past research in which Tullman-Ercek and collaborators developed a new technique, called SyMAPS (Systematic Mutation and Assembled Particle Selection), to test variations of a protein used by a bacterial virus called the MS2 bacteriophage. By substituting amino acids one at a time along the MS2 protein chain, the team could study how the virus scaffold was impacted by the different combinations, including which changes preserved the shell’s structure or broke it apart.
The latest research builds on the team’s progress by using SyMAPS to analyze multiple amino acid changes within the MS2 particle, a requirement to effectively manipulate virus shells in the future, Tullman-Ercek said. Researchers studied every double amino acid combination along a polypeptide loop located within the MS2 scaffold and measured how the virus scaffold was affected.
One factor producing epistasis was balancing the amino acid charges that were substituted, said Tullman-Ercek, a member of Northwestern’s Center for Synthetic Biology. Swapping two positively charged amino acids, for instance, caused the particle to repel and break apart, but balancing a single positive amino acid change with a separate negative charge compensated the switch and preserved stability.
“It looked like an unpredictable effect, but if you look at the overall trends of the data, we learned that charge is really important to maintain balance,” Tullman-Ercek said. “We couldn’t see that based on the data we accumulated with the single changes, but we kept coming back to this charge balancing issue.”
The team plans to expand testing to determine if the behaviors found in the MS2 particle apply to similar viruses.
“It takes years to optimize each component of a virus scaffold,” Tullman-Ercek said. “We’re trying to reduce the time it takes to optimize the delivery vehicle by learning the rules of how it assembles so we can eventually build one from scratch.”
Depending on its purpose or final destination in the body, virus scaffolds require unique design properties. A virus deployed to the brain to treat a tumor, for example, may need greater stability in its shape than one sent to the lungs. The more general the rules governing design, the greater variety of particles can be constructed and deployed in the future.
“If we have to optimize the delivery vehicle for every individual case, it will take decades to make progress, so figuring out the underlying rules is important,” she said. “It’s a fundamental science project, but it has the potential to really impact the design of a lot of future therapies.”
The insights also prompted the team to question how their strategy can be coupled with what is already known — and unknown — about evolution.
“In evolution, changes build on each other one at a time,” Tullman-Ercek said. “We’re making these changes deliberately in our lab, which makes you wonder how nature reaches these epistatic states with combinations that would produce negative results on their own. We want to build this for drug delivery, but the results raise interesting questions about how changes are optimized in nature to begin with.”