New Technique Offers Higher Resolution Molecular Imaging and Analysis
Approach could help researchers understand complicated biomolecular interactions and characterize diseases at the single-molecule level
A Northwestern Engineering research team has developed a new method to conduct spectroscopic nanoscopy, an approach that could help researchers understand more complicated biomolecular interactions and characterize cells and diseases at the single-molecule level.
The new system, called asymmetrically dispersed spectroscopic single-molecule localization microscopy (SDsSMLM), builds upon existing sSMLM techniques developed at the McCormick School of Engineering to provide more precise spectroscopic single-molecule analysis to study how cells behind certain cancers, or diseases like diabetic retinopathy, function in their localized environments.
 While current spectroscopic single-molecule localization microscopy techniques achieve super-resolution imaging and single-molecule spectroscopy simultaneously, current sSMLM designs suffer from reduced imaging resolution and spectral precision. This is caused by the system dividing a finite number of emitted photons — atomic particles that transmit electromagnetic light — between two separate channels for spatial and spectral imaging.
While current spectroscopic single-molecule localization microscopy techniques achieve super-resolution imaging and single-molecule spectroscopy simultaneously, current sSMLM designs suffer from reduced imaging resolution and spectral precision. This is caused by the system dividing a finite number of emitted photons — atomic particles that transmit electromagnetic light — between two separate channels for spatial and spectral imaging.
“We should not be satisfied to only know where a particular molecule is or where many molecules are without differentiating their properties,” said Hao Zhang, professor of biomedical engineering, who led the research. “Our approach enables us to fully use all photons from each emission for both spatial imaging and spectral analyses. As a result, we significantly improved the spatial imaging resolution and spectral precision compared to existing sSMLM techniques.”
A paper describing the work, titled “Symmetrically Dispersed Spectroscopic Single-molecule
Localization Microscopy,” was published May 25 in the journal Light: Science and Applications. Cheng Sun, professor of mechanical engineering, was a coauthor on the paper.
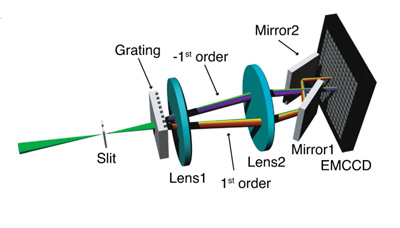
 Unlike existing sSMLM approaches, which often use a 1:3 ratio to split photos between the spatial and spectral channels, SDsSMLM commits all available photos to create two mirrored spectral images. This approach extracts spectral information at the highest possible resolution. In addition, because the images are symmetrical, researchers can still identify spatial information by identifying the mid-point between the two spectral images.
Unlike existing sSMLM approaches, which often use a 1:3 ratio to split photos between the spatial and spectral channels, SDsSMLM commits all available photos to create two mirrored spectral images. This approach extracts spectral information at the highest possible resolution. In addition, because the images are symmetrical, researchers can still identify spatial information by identifying the mid-point between the two spectral images.
When compared to an existing sSMLM using the same number of photos, the researchers found that SDsSMLM improved the spatial precision by 42 percent and spectral precision by 10 percent.
“We realized that the spatial information is totally overlooked in the spectral image in existing sSMLM techniques,” Zhang said. “This approach allows us to apply all of the available photons for spectral analysis to push the resolution limit while also acquiring spatial imaging.”
When used together with spectroscopic single-molecule imaging techniques, SDsSMLM can be adapted for 3D cellular imaging, an essential tool in cell biology and materials science that allows researchers to track how cells interact in their environments.
 “This technique holds true for all molecules, regardless of their emission spectra and minute spectral variations, even among the same species of molecules,” Zhang said. “With improved spatial resolution and spectral precision, sSMLM will find broader applications in multi-molecule imaging in cells and three-dimensional tracking for individual nanoparticles in biological and chemistry investigations.”
“This technique holds true for all molecules, regardless of their emission spectra and minute spectral variations, even among the same species of molecules,” Zhang said. “With improved spatial resolution and spectral precision, sSMLM will find broader applications in multi-molecule imaging in cells and three-dimensional tracking for individual nanoparticles in biological and chemistry investigations.”
In addition to the system’s advanced imaging capabilities, SDsSMLM’s compact nature allows for easy integration and reliable operation with conventional fluorescence microscope systems. Combined with an open source plug-in the researchers developed called RainbowSTORM, Zhang hopes other members of the biological research community will incorporate this advanced technique within their own work.
“Our design is standalone and can be installed in a majority of microscope systems,” Zhang said. “We hope other researchers take advantage of what we’ve created.”