Engineering Biosensors to Monitor Metabolism in Living Cells
Generalizable approach could enable the construction of biosensors for a wide range of metabolites
Northwestern Engineering’s Joshua N. Leonard has developed a new method for engineering novel biosensors that can monitor a cell’s inner state.
Leonard’s generalizable approach could potentially enable the construction of a wide range of biosensors for different purposes, including regulating a cell’s metabolic processes to drive it to produce desired products for pharmaceuticals, energy, and other biotechnology applications.
 “A lot of times, we might know what a cell is theoretically able to do, but we don’t have a way to force it to do what we want,” said Leonard, associate professor of chemical and biological engineering in Northwestern’s McCormick School of Engineering. “From a metabolic engineering perspective, it would be nice to have a tool that allows us to sense what’s happening inside a cell and then push its metabolism toward the production of things that we want.”
“A lot of times, we might know what a cell is theoretically able to do, but we don’t have a way to force it to do what we want,” said Leonard, associate professor of chemical and biological engineering in Northwestern’s McCormick School of Engineering. “From a metabolic engineering perspective, it would be nice to have a tool that allows us to sense what’s happening inside a cell and then push its metabolism toward the production of things that we want.”
Cells naturally use many internal biosensors to balance the various biochemical transformations that make up their metabolism. These biosensors help the cell to evaluate, regulate, and balance the conversion of chemicals into products that the cell needs to survive. If synthetic biologists could harness and engineer such a sensing mechanism, they could potentially implement molecular-level process control, which is a strategy used to great effect in virtually every factory and chemical plant. This biosensing ability is currently one of the biggest unmet needs in the biotechnology industry.
“A handful of biosensors have evolved to serve cells’ purposes, and those biosensors are indeed useful ‘parts,’” Leonard said. “However, there are far more metabolites that we, as engineers, would like to sense and build systems around than the naturally evolved biosensors can detect.”
Supported by the National Science Foundation and the Environmental Protection Agency, the research was published in the journal ACS Synthetic Biology. Andrew K. D. Younger, a graduate student in Leonard’s laboratory, was the paper’s first author. A founding member of Northwestern’s Center for Synthetic Biology, Leonard is also co-director of the Biotechnology Training Program and a member of the Chemistry of Life Processes Institute and Robert H. Lurie Comprehensive Cancer Center.
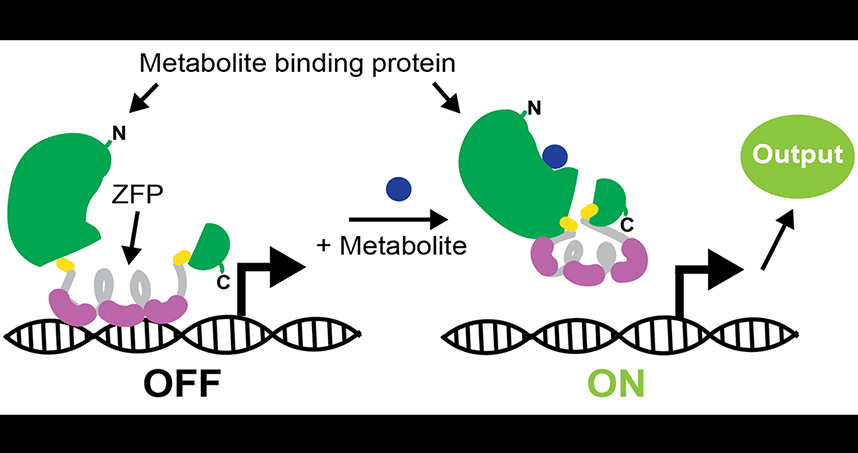
To develop and test their strategy for engineering new biosensors, Leonard’s team first used a well-studied protein that senses the presence of a metabolite — in this case a type of sugar called maltose — in the cell. Other research groups have fused this protein to a second fluorescent protein, such that the combined protein glows when the maltose is present, but this effect is transient and cannot be used to control cell state. To meet the needs described above, Leonard needed a biosensor that controls gene expression. In this study, Leonard’s team successfully developed a biosensor that drives expression of a reporter gene when maltose is present, providing a way to “record” the presence of maltose and to effectively change the cell’s state in response to maltose.
Leonard and Younger view this project as a starting point that could ultimately enable them and others to convert many types of metabolite-binding proteins into biosensors.
“What’s exciting is that our biosensor demonstrates a conversion strategy that is potentially generalizable,” said Younger, a graduate student in Northwestern’s Interdisciplinary Biological Sciences Graduate Program. “There is potentially nothing that we would need to change about our method to build biosensors from any other metabolite-binding protein.”
Someday, these biosensors could enable scientists to isolate rare and desirable genetic variants that produce metabolites of interest, perhaps from within mixed collections of genetically diverse cells.
By recording the presence of a desired metabolite via driving the production of a fluorescent reporter protein, biosensors could enable one to screen very large libraries, which remains beyond the capacity of even contemporary automated platforms.
“If you have a huge number of cells, but only one is making the metabolite that you want, it’s easy to pull out an individual cell if it’s fluorescent,” Leonard said. “A biosensor like this allows you to find the needle in the haystack.”